September 16, 2019 - by Sarah Waldrip
If you’ve ever suffered from heartburn or gastric ulcers, then you know first-hand how gastric acid can wreak havoc when it is too abundant in the body. In order to learn more about the complex physiological process that produces it, Himanshu Khandelia, Associate Professor at the University of Southern Denmark, collaborated with a team of scientists in Japan to use the “Piz Daint” supercomputer at the Swiss National Supercomputing Center (CSCS) to run molecular dynamics (MD) simulations of a pivotal moment in gastric acid production in the interior stomach lining.
These simulations helped finally resolve a long-running scientific debate by showing the precise rate of ion exchange, or “stoichiometry”, required to produce so much acid, as well as how highly specialized enzymes manage the job on a structural level. Khandelia’s research team recently completed their year-long study and published their findings in the scientific journal eLIFE.
Digesting the Facts
The concoction of juices in the human stomach, known collectively as gastric acid, is composed of two relatively neutral compounds, potassium chloride and sodium chloride, but also highly corrosive hydrochloric acid (HCl). With a pH of 1, hydrochloric acid is nearly as corrosive as battery acid. It’s so acidic that it can cause burns when it comes in contact with unprotected skin, and it’s powerful enough to strip paint from a surface and even dissolve many types of metal. On a molecular level, acids of this strength also are known to be very rich in positively charged hydrogen ions (hydrogen atoms with extra protons), which is a key element in gastric acid production.
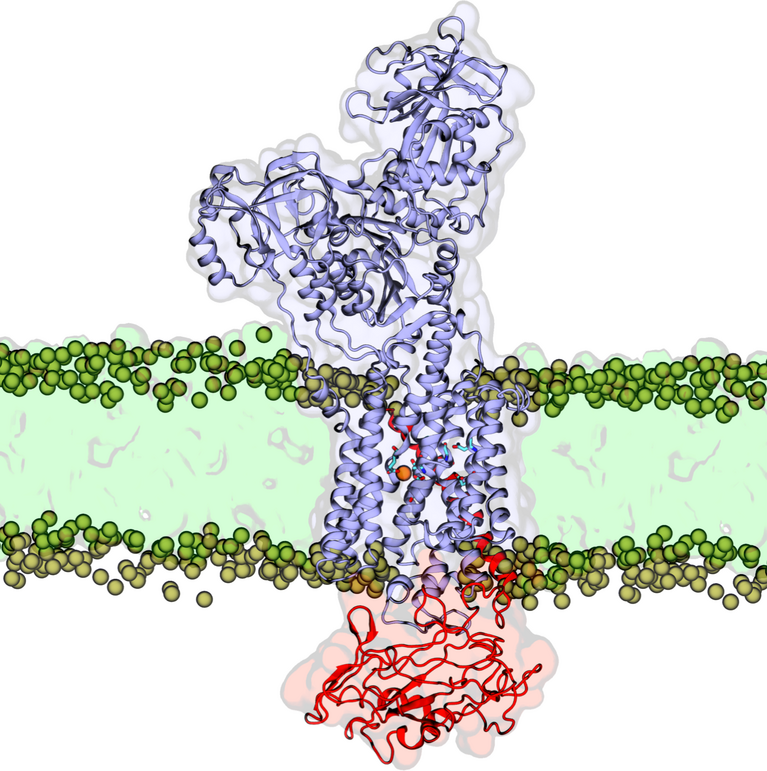
Whereas hydrochloric acid alone has a pH level of 1, the blend of secretions in the stomach is normally between pH 1.5 and 3—acidic enough to digest food and fight infection effectively, but not so acidic that it harms the body. This delicate balance is largely maintained by several specialized groups of cells in the lining of the stomach. However, it was a molecular process in one particular type of cell, the parietal cell, that Khandelia and his fellow researchers have been studying for the past year. Specifically, they sought to determine how one particular enzyme raises acidity in the stomach by interacting with the parietal cells.
This tiny enzyme, gastric hydrogen potassium ATPase, or simply called the gastric proton pump, accomplishes the incredible feat of maintaining such high acidity by pumping positively charged hydrogen ions (H+) back into the already-hydrogen-rich acid in the stomach in exchange for the more scarce potassium ions (K+), which are counter-transported into the cell’s pH-neutral, electro-neutral cytoplasm.
This laborious process results in the secretion of gastric acid at an extremely high proton gradient across the cell membrane—meaning, the gastric proton pump is pushing against an enormous amount of resistance to force H+ across the cell membrane and back into the stomach. Because the already-present gastric acid is so strong, the pump is pushing back against a million-fold H+ gradient in fact, “one of the highest cation gradients known in mammalian tissue”, write the scientists in their publication. The “cation” in this case is the positively charged ion that is being transported across the membrane. This process must be completed for new gastric acid to be secreted. And as the scientists also noted, “The secretion of a relatively voluminous flow of gastric acid requires the expenditure of considerable cellular energy.” So how does the gastric proton pump manage all of this?
Learning from Molecular Dynamics Simulations
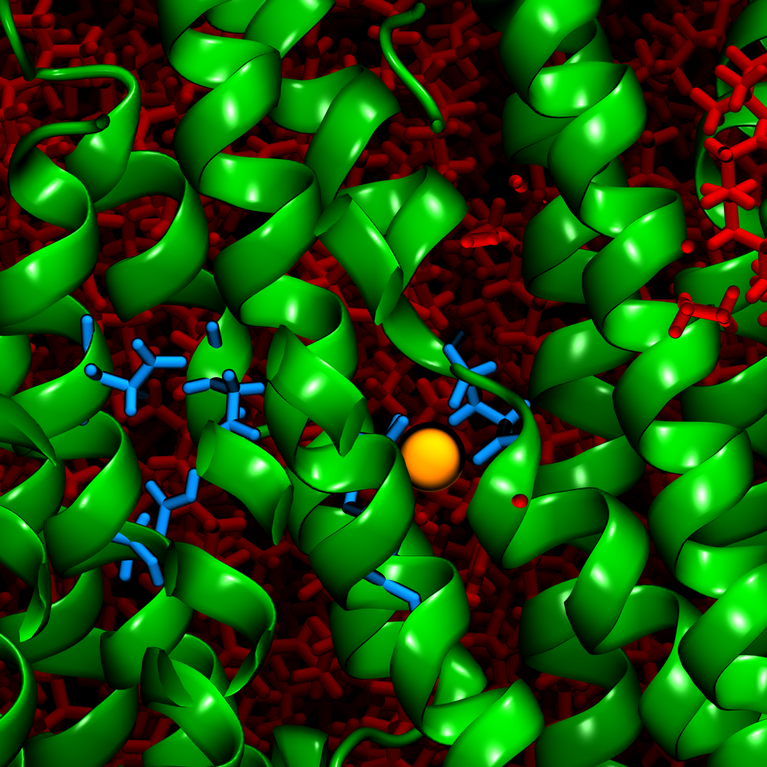
The major findings for Khandelia and his team came when their carefully planned simulations revealed the stoichiometry, i.e. exactly how many protons the gastric proton pump could exchange for the cost of one unit of cellular energy (one ATP), and also how the proton pump was able to achieve this exchange on a structural level. What they found was that the proton exchange was expensive—one ATP of energy was only enough to exchange one K+ for one H+ given the conditions. Further, the enzyme pump was found to contain a unique crystal structure that controlled this ion exchange through a “luminal gate closure”. This “gate” serves as a mechanism to block any extra H+ from binding to the transport site, despite its abundance, and instead to allow only a single K+ to be transported within the enzyme.
In order to watch the precise moment these ions interact with the gastric proton pump, researchers simulated several simulation configurations of approximately 350,000 atoms for a total time span of 12 microseconds. This required 120,000 hours of compute nodes on the “Piz Daint” supercomputer at CSCS.
These findings would have been practically impossible to achieve without the molecular dynamics (MD) simulations created with “Piz Daint”, according to Khandelia. “In general, for us, MD simulations are a tool which we use to explore the dynamics of macromolecules and interactions on a sub-nanometer scale, which is not possible to do using most other experimental techniques,” he says. “All of my research has employed MD simulations.”
Revealing a Path Forward
Solving puzzles like the gastric proton pump will ripple through the bioscience research fields. “This work can spawn similar research into a wide variety of active membrane proteins, which pump not only ions, but also drugs out of the cells in bacteria, for example,” says Khandelia. “These findings can, for example, provide the basis for rational design of new drugs which treat problems such as chronic acidity and stomach ulcers.” With this research now concluded, the research team is already moving on to other related studies. “Our own group is now working on a protein which is involved with transporting cholesterol across the membrane,” Khandelia notes.
In any case, the way forward for this research is likely to remain highly dependent on high-performance computing (HPC). “We cannot pursue our research without HPC,” says Khandelia. “For us, it is like water in the desert.”
Reference:
Yamamoto K, Dubey V, Irie K, Nakanishi H, Khandelia H, Fujiyoshi Y & Abe K: A single K+-binding site in the crystal structure of the gastric proton pump, eLife August 2019, Doi: 107554/eLife.47701.
About the scientific work of Himanshu Khandelia and his group: https://himanshukhandelia.wordpress.com/about/